Table of contents
Introduction
What is statistics?
The field of statistics – the practice and study of collecting and analyzing data.
A summary statistic – a fact about or summary of some data, like an average or a count.
What can statistics do?
With the power of statistics, we can answer tons of different questions like:
How likely is someone to purchase a product? Are people more likely to purchase it if they can use a different payment system?
How many occupants will your hotel have? How can you optimize occupancy?
A/B Testing: Which ad campaign is more effective in getting people to purchase a product?
What can't statistics do?
Why is One Piece so popular?
Could not use to find out why a relationship exists
Types of statistics
| Descriptive Statistics | Inferential Statistics |
| Describe and summarize the data | Use a sample of data to make inferences about a larger population |
Types of data
Numeric (Quantitative)
Continuous (Measured)
Example
Car speed
Time spend waiting in line
Discrete (Counted)
Example
Number of pets
Number of packages shipped
Categorical (Qualitative)
Nominal (Unordered)
Example
Country of residence
Male / Female
Ordinal (Ordered)
Example
Strongly disagree
Somewhat disagree
Neutral
Somewhat agree
Strongly agree
Categorical data can be represented as numbers
Nominal (unordered)
Male / Female (
1,2)Country of residence (
1,2, ...)
Ordinal (ordered)
Strongly disagree (
1)Somewhat disagree (
2)Neutral (
3)Strongly agree (
4)Somewhat agree (
5)
Why does data types matter?
The type of data will dictate what kinds of summary statistics and visualizations make sense for our data.
For numerical, we can use summary statistics like mean, and plots like scatter plots, but this doesn't make sense for categorical data. Similarly, counts and bar plots don't make much sense for numeric data
Measures of the center
Where is the center of the data?
Mean
mean = np.mean(data_frame['column'])Median
median = np.median(data_frame['column'])Mode (most frequent value of the data)
data_frame['column'].value_counts()
Which one to use?
We have lots of ways to measure center.
Mean
Mean works better for symmetrical data
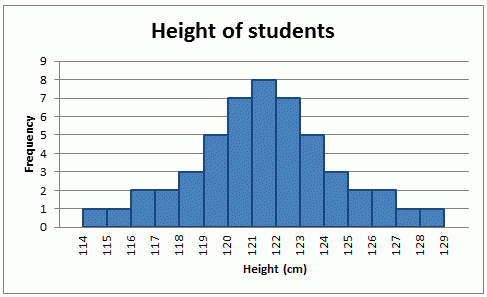
Median
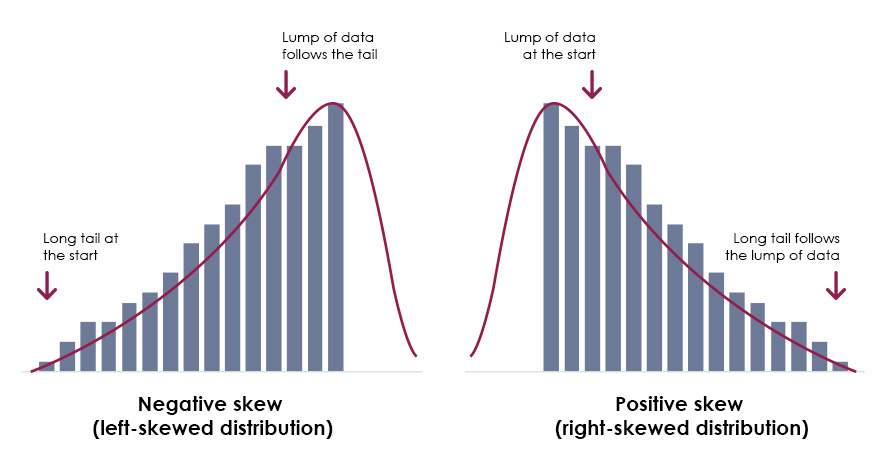
Median is usually better to use for skewed data. Because the mean is pulled around by the extreme values, it's better to use the median since it's less affected by outliers.
Mean and median
# Import numpy with alias np
import numpy as np
# Filter for Belgium
be_consumption = food_consumption[food_consumption['country'] == 'Belgium']
# Filter for USA
usa_consumption = food_consumption[food_consumption['country'] == 'USA']
# Calculate mean and median consumption in Belgium
print(np.mean(be_consumption['consumption']))
print(np.median(be_consumption['consumption']))
# Calculate mean and median consumption in USA
print(np.mean(usa_consumption['consumption']))
print(np.median(usa_consumption['consumption']))
# Import numpy as np
import numpy as np
# Subset for Belgium and USA only
be_and_usa = food_consumption[food_consumption['country'].isin(['Belgium', 'USA'])]
# Group by country, select consumption column, and compute mean and median
print(be_and_usa.groupby('country')['consumption'].agg([np.mean, np.median]))
# Import matplotlib.pyplot with alias plt
import matplotlib.pyplot as plt
# Subset for food_category equals rice
rice_consumption = food_consumption[food_consumption['food_category'] == 'rice']
# Histogram of co2_emission for rice and show plot
rice_consumption['co2_emission'].hist()
plt.show()
# Subset for food_category equals rice
rice_consumption = food_consumption[food_consumption['food_category'] == 'rice']
# Calculate mean and median of co2_emission with .agg()
print(rice_consumption.agg([np.mean, np.median]))
Measures of spread
What is spread?
Describes how spread apart or close together the data points are. There are a few different measures of spread.
Variance, The average distance from each data point to the data's mean. The higher the variance the more spread out the data is.
# 1. Calculating distance between each point and the mean distances = msleep['sleep_total'] - np.mean(msleep['sleep_total']) # 2. Square each distance squared_distances = distances ** 2 # 3. Sum squared distance sum_squared_distances = np.sum(squared_distances) # 4. Divide by number of data points - 1 variance = sum_squared_distance / len(squared_distance) - 1 # Otherwise variance = np.var(msleep['sleep_total'], ddof=1)ddof: “Delta Degrees of Freedom”: the divisor used in the calculation is
N - ddof, whereNrepresents the number of elements. By default ddof is zero.Standard Deviation is a measure of the amount of the variation of set of values.
Low standard deviation indicates that the values tend to be closed to the mean (also called the expected value) of the set
High standard deviation indicates that the values are spread out over a wider range
std = np.sqrt(np.var(msleep['sleep_total'], ddof=1))
# Otherwise
std = np.std(msleep['sleep_total'], ddof=1)
Mean absolute deviation is the average distance between each data point and the mean.
standard deviation vs mean absolute deviation
Standard deviation squares distance, penalizing longer distances more than shorter ones. Mean absolute deviation penalizes distances equally.
One isn't better than others, but SD more common than MAD.
distances = msleep['sleep_total'] - np.mean(msleep['sleep_total'])
mad = np.mean(np.abs(distances))
Quantiles are cut points dividing the range of a probability distribution into continuous intervals with equal probability. In other words we can say that quantiles split up the data into some number of equal part
median = np.quantile(msleep['sleep_total'], 0.5) quartiles = np.quantile(msleep['sleep_total'], [0, 0.25, 0.5, 0.75, 1]) # Using linspace quartiles = np.quantile(msleep['sleep_total'], np.linespace(0, 1, 5))Boxplots use quartiles

Interquartile range (IQR) is the distance between 25th and 75th percentile, which is also height of the boxplot
iqr_result = np.quantile(msleep['sleep_total'], 0.75) - np.quantile(msleep['sleep_total'], 0.25) # Using scipy stats from scipy.stats import iqr iqr_result = iqr(msleep['sleep_total'])Outliers: data point that is substantially different from others.
from scipy.stats import iqr iqr_result = iqr(msleep['sleep_total']) lower_threshold = np.quantile(msleep['sleep_total'], 0.25) - 1.5 * iqr_result upper_threshold = np.quantile(msleep['sleep_total'], 0.75) + 1.5 * iqr_result
There are many statistics we've covered earlier, we can get all statitics in just one line of code using describe method. It's convinient to use when we want to get a general sense of our data.
msleep['sleep_total'].describe()
Quartiles, quantiles, and quintiles
# Calculate the quartiles of co2_emission
print(np.quantile(food_consumption['co2_emission'], np.linspace(0, 1, 5)))
# Calculate the quintiles of co2_emission into 5 pieces
print(np.quantile(food_consumption['co2_emission'], np.linspace(0, 1, 6)))
# Calculate the deciles of co2_emission
# print([x x in 0.10])
print(np.quantile(food_consumption['co2_emission'], np.linspace(0, 1, 11)))
Variance and standard deviation
# Print variance and sd of co2_emission for each food_category
print(food_consumption.groupby('food_category')['co2_emission'].agg([np.var, np.std]))
# Import matplotlib.pyplot with alias plt
import matplotlib.pyplot as plt
# Create histogram of co2_emission for food_category 'beef'
food_consumption[food_consumption['food_category'] == 'beef']['co2_emission'].hist()
# Show plot
plt.show()
# Create histogram of co2_emission for food_category 'eggs'
food_consumption[food_consumption['food_category'] == 'eggs']['co2_emission'].hist()
# Show plot
plt.show()
Finding outliers using IQR
# Calculate total co2_emission per country: emissions_by_country
emissions_by_country = food_consumption.groupby('country')['co2_emission'].sum()
print(emissions_by_country)
# Compute the first and third quartiles and IQR of emissions_by_country
q1, q3 = np.quantile(emissions_by_country, [0.25, 0.75])
iqr = q3 - q1
# Calculate the lower and upper cutoffs for outliers
lower = q1 - 1.5 * iqr
upper = q3 + 1.5 * iqr
# Subset emissions_by_country to find outliers
outliers = emissions_by_country[(emissions_by_country < lower) | (emissions_by_country > upper)]
print(outliers)

